| |
Human herpesvirus detection and species
identification with PCR-RFLP and ELISA: a
comparative study
Guan-Ping Dong, Shi-Qiang Shang, Li-Zhong Du, Xi-Lin Yu, Ya-Ping Xu, Xiu-Jing Wu
Hangzhou, China
Author Affiliations: Department of Infection, Children's Hospital, Zhejiang University School of Medicine, Hangzhou 310003, China (Dong GP, Shang SQ, Du LZ, Yu XL, Xu YP, Wu XJ)
Corresponding Author: Guan-Ping Dong, MD, Department of Infection, Children's Hospital of Zhejiang University School of Medicine, Hangzhou 310003, China (Tel: 86-571-87061007 ext 61412; Fax: 86-571-87033296; Email: dongguanp@yahoo.com.cn)
Background: This study was undertaken to establish a restriction endonulease pattern which could detect and differentiate four major human herpesviruses by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP), and to compare PCR-RFLP with enzyme-linked immunosorbent assay (ELISA) in diagnosing herpesvirus infection.
Methods: A pair of primers was designed to amplify herpes simplex virus type 1 and 2 (HSV-1/-2), Epstein-Barr virus (EBV) and cytomegalovirus (CMV). At last, we used the PCR-RFLP technique to differentiate four different herpesviruses. Meanwhile, 75 clinical blood specimens from infants of suspected viral infection and 38 blood specimens from healthy children were evaluated for herpesviruses DNA by PCR-RFLP or virus-specific IgM antibody detection by ELISA.
Results: The products of four human herpesviruses after PCR amplification varied from 510 bp to 592 bp and allowed characterization of herpesvirus type with restriction endonulease analysis. Among the 75 specimens, 23 (30.7%) were shown positive by PCR including 13 for CMV, 4 for EBV, 5 for HSV-2, and 1 for HSV-1 after restriction enzyme digestion with BamHI and BstUI, whereas 10 (13.3%) were detected positive by ELISA. All ELISA-positive specimens were likewise positive by PCR. Thirteen of 65 ELISA-negative specimens were tested positive by PCR.
Conclusion: The PCR-RFLP technique is more specific, sensitive, rapid and accurate than ELISA in diagnosing herpesvirus infection.
Key words: herpesvirus; polymerase chain reaction; restriction fragment length; polymorphism; ELISA
World J Pediatr 2007;3(2):129-133
Introduction
Human herpesviruses including herpes simplex virus type 1 (HSV-1), type 2 (HSV-2), cytomegalovirus (CMV) and Epstein-Barr virus (EBV) commonly cause infections of the central nervous system and urinary tract. Clinical signs and symptoms resulting from infections of these four different viruses are indistinguishable.[1,2] Establishing a specific viral diagnosis is now essential for a specific anti-viral therapy.
In our study, a pair of primers was designed according to the well-conserved region of DNA polymerase gene of the four herpesviruses. With this pair of primers and with BamHI, BstUI endonucleases, the polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) technique was performed to identify the four herpesviruses. Finally, the PCR method was evaluated in comparison with enzyme-linked immunosorbent assay (ELISA) on blood specimens of children referred to our laboratory from June 2001 to December 2002.
Methods
Clinical specimens
From June 2001 to December 2002, blood specimens from 75 infants suspected of viral infection at Children's Hospital of Zhejiang University, China were evaluated by PCR-RFLP and ELISA. In addition, blood specimens from 38 healthy children were used as controls and also evaluated by PCR and ELISA.
Blood specimens were obtained from each patient within 24 hours of hospitalization. Four milliliters venous blood was taken and separated into 2 ml per tube. One is anticoagulated for DNA isolation and the other coagulated for serum isolation. All of the specimens were stored at -70ºC until use.
Reference virus strains
Reference virus strains CMV (strain AD169, cultivated in human fibroblast cells), VZV (strain EF, cultivated in vero cells), HSV-2 and HSV-1 (strain G and strain F, propagated in a vero cell line) were provided by Department of Microbiology, Anhui Medical University, Anhui, China. They were used as positive controls for amplification with herpesvirus consensus primers. E. coil, S. aureus, hepatitis B virus (HBV), C. neoformans and human-genomic DNA obtained from Children's Hospital of Zhejiang University were used as negative controls.
DNA isolation
All virus-infected cell cultures and blood specimens were treated with 300 µl of lysis buffer including 0.2 mmol Tris-HCl (pH7.5), 25 mmol EDTA, 0.3 mmol NaCl, 2% SDS, and 400 mg/L proteinase K (Takara Biotechnology, DALIAN Co. Ltd, China). The mixture was incubated at 56ºC for 2 hours and then extracted with phenol-chloroform-isoamyl alcohol (25/24/1) before PCR. The DNA was precipitated with absolute alcohol. The DNA pellets were washed with 70% alcohol, dried and then suspended in 20 µl of sterile distilled water. Blood specimens in the presence of heparin were subjected to red cell lysis in five volumes of 0.87% NH4Cl, and leukocytes were pelleted and suspended in 500 µl of this lysis buffer. Finally, the resulting DNA preparations from blood specimens were stored at -70ºC.
Amplification with herpesvirus consensus primers
Oligonucleotide primers were designed to bracket a well-conserved region in the DNA polymerase gene, based on an alignment of the DNA sequences of the four known human herpesviruses through Genbank. Sequences of primers P1 (5'-CGACTTTGCCAGCCTGACC-3') and P2 (5'-AGTCCGTGTCCCCGTAGATG-3') were used to amplify HSV-1, HSV-2, EBV and CMV.
PCR was done in 50 µl reaction mixture containing 5 µl of 10 ¡Á buffer, 5 µl of 25 mmol MgCl2, 4 µl of a deoxyucleoside triphosphate mixture (each deoxyucleoside triphosphateat concentration of 2.5 mmol), 2.5 µl of dimethyl sulfoxide, 50 pmol of each oligonucleotide primer, 0.5 µl (2.5 U) of Taq DNA polymerase (Takara Biotechnology, DALIAN Co. Ltd, China), 15 µl of extracted specimen DNA, and double-distilled water to a volume of 50 µl. The reaction mixture was overlaid with mineral oil (Takara Biotechnology, DALIAN Co. Ltd, China). The cycling parameters of PCR were initial preincubation at 95ºC for 5 minutes and then 36 cycles consisting of 95ºC for 1 minute, 55ºC for 1 minute, and 72ºC for 1 minute, followed by a final incubation at 72ºC for 5 minutes on a 9600 Research thermocycler (Perkin-Elmer, Norwalk, Conn). A 10 µl volume of each reaction mixture was analyzed by electrophoresis on 1.5% agarose gels containing ethidium bromide using a UV transilluminator and then examined and photographed. Negative and positive controls were run in each experiment.
Sensitivity of PCR
The sensitivity of primers was determined with reference strains including CMV, EBV, HSV-1, and HSV-2. The DNA extracted from tissue culture bottles was diluted serially in 10-fold dilutions (range from 100 pg to 0.01 fg) and then subjected to PCR. The extracted DNA was read spectrophotometrically (Beckman DU 640, USA) at 260 nm for the quantitation of the virus. The sensitivity of primers with reference strains was thus calculated.
Restriction enzyme analysis
Each reaction mixture in which amplicons were detected was subjected to digestion with restriction enzymes BamHI (Takara) and BstUI (MBI). The digestion mixture consisted of 10 µl of PCR mixture, 2 µl of appropriate enzyme buffer, 1.5 µl (15 U) of enzyme and 6.5 µl of double-distilled water for a total volume of 20 µl. The reaction mixture was incubated for 2 hours at 37ºC. The digested PCR products were resolved on 2.5% agarose gels containing ethidium bromide. The gels were visualized and photographed using a UV transilluminator. The DNA fragment sizes were compared to a DNA molecular weight marker (2000 bases DNA Ladder, Takara, USA).
Herpesviral-specific IgM antibody detection
The ELISA kits (EUROIMMUN, Germany) were used to determine viral-specific IgM antibody from 75 blood specimens according to the manufacturer's protocol.
Results
Sensitivity and specificity of the PCR assay
The generation of PCR products of the expected size (592 bp for CMV, 520 bp for EBV, 518 bp for HSV-1/2) attested to the specificity of the PCR assay. These results demonstrated that there was no cross-reaction to the control specimens and the assay was highly specific for the herpesviruses (Fig. 1). To determine the limits of viral detection, the sensitivity of the PCR assay with the pair of primers was assessed with serial dilutions of DNA with specified concentrations (range 100 pg to 0.01 fg) and added to the reaction mixture. The limiting sensitivities measured were 1 fg DNA for HSV-2 and 0.1 fg DNA for CMV, EBV and HSV-1.
Identification of herpesviruses species
According to the sequencing of the 4 human herpesviruses, we noted that amplicons obtained by PCR from the template DNAs described above were subjected to restriction endonuclease digestion with BamHI and BstUI. The CMV amplicon of 592 bp was cleaved by BstUI into nine fragments (170 bp, 130 bp, 80 bp, 62 bp, 50 bp, 40 bp, 30 bp, 16 bp and 14 bp) and remained undigested by BamHI. The EBV amplicon of 520 bp cleaved by BamHI and BstUI into two fragments (250 bp and 270 bp, 254 bp and 266 bp, respectively). The HSV-1 amplicon of 518 bp was cleaved by BstUI into eight fragments (230 bp, 130 bp, 50 bp, 40 bp, 23 bp, 20 bp, 15 bp and 10 bp) and remained undigested by BamHI. The HSV-2 amplicon of 518 bp was cleaved by BamHI into two fragments (230 bp and 288 bp) and by BstUI into ten fragments (168 bp, 90 bp, 80 bp, 60 bp, 40 bp, 30 bp, 20 bp, 15 bp, 10 bp and 5 bp). After digestion and electrophoresis, each viral DNA yielded a specific fragment pattern that could be differentiated from each other (Fig. 2).
Testing of clinical specimens
The clinical characteristics of the patients who tested positive by the two tests are presented in the Table. Clinical specimens from patients hospitalized for about one year period were tested for herpesvirus DNA by PCR assay or viral-specific IgM antibody by ELISA. Among the 75 specimens, 23 (30.7%) were shown positive by PCR. They included CMV (13), EBV (4), HSV-2 (5) and HSV-1 (1) after restriction enzyme digestion with BamHI and BstUI, whereas 10 (13.3%) were shown positive by ELISA. All ELISA-positive specimens were shown likewise positive by PCR. Thirteen of 65 specimens that were ELISA-negative were tested positive by PCR. An infant with CMV infection was determined viral DNA and virus-specific IgM antibody in blood at 3, 4 and 6 months after birth, respectively. None of the 38 control blood specimens was positive for the herpesvirus by PCR-RFLP or ELISA.
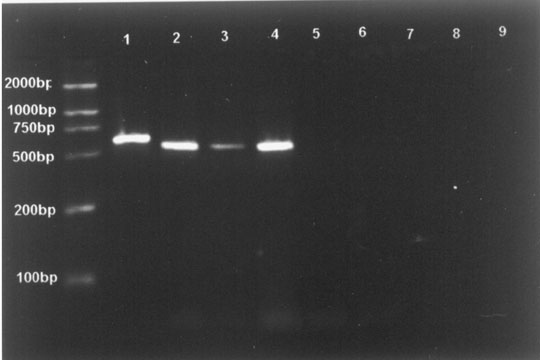 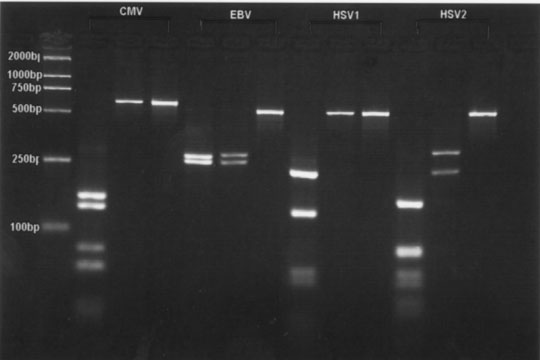
Fig.1. Fig.2.
Fig. 1. Specificity of PCR amplification. Lane M: molecular weight standards (2000 bases DNA Ladder); Lane 1: CMV; Lane 2: EBV; Lane 3: HSV-1; Lane 4: HSV-2; Lane 5: E. coil; Lane 6: S. aureus; Lane 7: hepatitis B virus; Lane 8: C. neoformans; Lane 9: human-genomic DNA. Fig. 2. Detection of CMV, EBV, HSV-1 and HSV-2 by restriction enzyme digest. Lane M: molecular weight standards (2000 bases DNA Ladder); Lane 1: fragments after digestion with BstUI; Lane 2: fragments after digestion with BamHI; Lane 3: amplified fragments without restriction enzyme analysis
Table. Clinical characteristics of the patients who were tested positive by PCR-RFLP and ELISA
|
No. Age (d) |
Sex (M/F) |
Symptoms* |
PCR |
RFLP |
IgM |
|
1 |
2 |
M |
1, 2, 3 |
+ |
HSV-2 |
(-) |
|
2 |
12 |
M |
2, 3, 4, 5 |
+ |
CMV |
CMV (+) |
|
3 |
240 |
F |
6 |
+ |
CMV |
(-) |
|
4 |
180 |
M |
7 |
+ |
CMV |
(-) |
|
5 |
150 |
M |
1, 8, 9 |
+ |
EBV |
EBV (+) |
|
6† |
90 |
F |
1, 7 |
+ |
CMV |
CMV (+) |
|
7 |
47 |
F |
1, 10 |
+ |
CMV |
CMV (+) |
|
8 |
60 |
M |
10, 11 |
+ |
EBV |
(-) |
|
9 |
3 |
M |
12, 13 |
+ |
HSV-2 |
(-) |
|
10 |
1 |
F |
14, 5 |
+ |
CMV |
(-) |
|
11 |
1 |
F |
2, 15 |
+ |
CMV |
(-) |
|
12 |
2 |
F |
1, 16 |
+ |
CMV |
(-) |
|
13 |
14 |
M |
2, 9 |
+ |
HSV-2 |
HSV-2 (+) |
|
14 |
15 |
M |
1, 17 |
+ |
EBV |
EBV (+) |
|
15 |
60 |
M |
5, 18 |
+ |
CMV |
(-) |
|
16 |
40 |
M |
19 |
+ |
CMV |
CMV (+) |
|
17 |
210 |
F |
20 |
+ |
CMV |
CMV (+) |
|
18 |
1 |
F |
19 |
+ |
HSV-2 |
HSV-2 (+) |
|
19 |
2 |
F |
16, 1 |
+ |
HSV-2 |
(-) |
|
20 |
60 |
M |
5, 7 |
+ |
CMV |
(-) |
|
21 |
270 |
M |
8, 9 |
+ |
HSV-1 |
(-) |
|
22 |
20 |
M |
2, 1 |
+ |
CMV |
(-) |
|
23 |
50 |
F |
1, 8, 9 |
+ |
EBV |
EBV (+) |
*: 1, hepatosplenomegaly; 2, congenital heart disease; 3, anemia; 4, umbilical hernia; 5, pneumonia; 6, intellect hypoevolutism; 7, microcephaly; 8, febrile; 9, skin eruption; 10, hyperbilirubinemia; 11, 21-trisomy syndrome; 12, undescended testis; 13, apnea; 14, congenital laryngeal stridor; 15, mirror-image heart; 16, convulsion; 17, thrombocytopenia; 18, idiopathic thromobocytopenlc purpura; 19, biliary atresia; 20, infectious mononucleosis. †: An infant with CMV infection was determined viral DNA and virus-specific IgM antibody in blood at 3, 4 and 6 months after birth, respectively. The result showed that she was still DNA-positive in blood in contrast to IgM-positive only at 3 months after birth (Abbreviations: M: male; F: female; HSV: herpes simplex virus; CMV: cytomegalovirus; EBV: Epstein-Barr virus).
Discussion
Because of its high molecular sensitivity and efficiency, PCR for DNA amplification has become the "gold standard" diagnostic test for diseases such as herpetic encephalitis,[3] but this has been claimed as an inappropriate diagnosis because of the slow appearance. Several studies[4,5] have reported a multiplex PCR method to identify viral infection of the central nervous system. However, these methods combined with several different primers may decrease their sensitivity. Furthermore, these protocols require a nested amplification to detect and classify herpesviruses. Although single PCR with consensus primers combined with Southern blot hybridization has been developed,[6] it is unable to distinguish HSV-1 from HSV-2. Their sequences are highly conserved in this region and it is difficult to select virus-type specific probes. In our study, we designed a pair of primers to amplify the four viruses. These primers were located in the well-conserved regions of the DNA polymerse gene of the human herpesviruses. Sequence analysis and virus species identification were done through restriction enzyme digestion with BamHI and BstUI. Finally, we developed the PCR-RFLP technique to differentiate the four different herpesviruses.
Results of our study demonstrated that using a highly sensitive and specific procedure (PCR) was possible to detect as low as 0.1 fg DNA for CMV. Moreover, there was no cross-reaction to human DNA, E. coli, S. aureus, HBV, and C. neoformans. For the species identification, BamHI profiles for CMV and HSV-1 are very similar and can be a source of misidentification.[7] However, BstUI from two to ten cutting sites with the gene region analyzed provides distinct restriction profiles for all viruses under investigation. Even for amplicons of CMV, HSV-1 and HSV-2 spliced several times by BstUI, the large fragments are identifiable (Fig. 2). Thus, BamHI and BstUI endonucleases are chosen to identify all viral DNAs and also proved to perform well with purifying amplicons from the PCR mixture.
ELISA is commonly regarded as an effective technique to detect the herpesviruses in the clinical specimens from patients, but viral-specific IgM antibody peaks 7-14 days by ELISA. The IgM response disappears again after 1 month,[8] then, only a minority of HSV-2 patients may be positive for IgM early (<30 days) during their first episode.[9] In addition, the herpesvirus-specific IgM response also has cross-reaction to other viruses,[10] lowering the sensitivity of ELISA. In our study, all ELISA-positive specimens were tested positive by PCR. Thirteen of the ELISA-negative 65 specimens were tested positive by PCR. An infant with CMV infection was determined viral DNA and virus-specific IgM antibody in blood at 3, 4 and 6 months after birth, respectively, showing DNA-positive in blood in contrast to IgM-positive only at 3 months after birth. These results showed that PCR-RFLP could detect herpesvirus DNA in specimens which were shown negative by ELISA.
In summary, the PCR-RFLP test can amplify and identify four human herpesviruses through coupling with digestion using a panel of two restriction enzymes. This assay is more sensitive, cost-effective and efficient than ELISA for the analysis of blood specimens in infants.
Acknowledgements
The authors want to thank Fang Yongming and Jiang Wenzhi for DNA cloning and sequencing, Yu Zhongsheng for valued support and advice, and Xu Yaping for collection of clinical specimens.
Funding: This project was supported by a grant from the Bureau of Science and Technology of Zhejiang Province (No. 011103999).
Ethical approval: No.
Competing interest: No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Contributors: DGP wrote the first draft of this paper. All authors contributed to the intellectual content and approved the final version. DGP is the guarantor.
References
1 Studahl M, Bergstrom T, Ekeland-sjoberg K, Ricksten A. Detection of cytomegalovirus DNA in cerebrospinal fluid in immunocompetent patients as a sign of active infection. J Med Virol 1995;46:274-280.
2 Studahl M, Hagberg L, Rekabdar E, Bergstrom T. Herpesvirus DNA detection in cerebral spinal fluid: differences in clinical presentation between alpha-, beta-, and gamma-herpesviruses. Scand J Infect Dis 2000;32:237-248.
3 Mitchell PS, Espy MJ, Smith TF, Toal DR, Rys PN, Berbari EF, et al. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J Clin Microbiol 1997;35:2873-2877.
4 Pozo F, Tenorio A. Detection and typing of lymphotropic herpesviruses by multiplex polymerase chain reaction. J Virol Methods 1999;79:9-19.
5 Read SJ, Kurtz JB. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J Clin Microbiol 1999;37:1352-1355.
6 Yamamoto T, Nakamura Y. A single tube PCR assay for simultaneous amplification of HSV-1/-2, VZV, CMV, HHV-6A/-6B, and EBV DNAs in cerebrospinal fluid from patients with virus-related neurological diseases. J Neurovirol 2000;6:410-417.
7 Demkin VV, Kruglova AI, Nikolaeva NP, Yurchenko JV. Detection and species identification of four human herpesviruses using polymerase chain reaction coupled with restriction endonuclease analysis. J Virol Methods 2002;103:121-128.
8 Stensballe LG, Kofoed PE, Nante EJ, Sambo M, Jensen IP, Aaby P. Duration of secretory IgM and IgA antibodies to respiratory syncytial virus in a community study in Guinea-Bissau. Acta Peadiatr 2000;89:421-426.
9 Morrow R, Friedrich D. Performance of a novel test for IgM and IgG antibodies in subjects with culture-documented genital herpes simplex virus-1 or -2 infection. Clin Microbiol Infect 2006;12:463-469.
10 Klutts JS, Liao RS, Dunne WM Jr, Gronowski AM. Evaluation of a multiplexed bead assay for assessment of Epstein-Barr virus (EBV) immunologic status. J Clin Microbiol 2004;42:4996-5000.
Received May 31, 2006; Accepted after revision March 4, 2007
|

