|
Detection of minimal residual disease in pediatric
patients with acute lymphoblastic leukemia and
its prognostic significance
Yan Bai, Hui Yu, Wen Lin, Dong-Feng Zhou, Yan Xiao, Xiao-Yan Wu,
Run-Ming Jin, Hong-Bao Fei
Wuhan, China
Author Affiliations: Department of Pediatrics, Union Hospital, Huazhong University of Science and Technology, Wuhan 430022, China (Bai Y, Yu H, Lin W, Zhou DF, Xiao Y, Wu XY, Jin RM, Fei HB)
Corresponding Author: Yan Bai, MD, Department of Pediatrics, Union Hospital, Huazhong University of Science and Technology, Wuhan 430022, China (Tel: 86-27-85726002; Fax: 86-27-85726002; Email: swcathy32@yahoo.com.cn)
Background: At diagnosis, leukemic cell burden reaches 1012 in patients with acute lymphoblastic leukemia (ALL). When patients get the first complete remission (CR) after induction chemotherapy, less than 5% of lymphoblasts are found in bone marrow smear by light microscopy. But an amount of 109 or less residual leukemic cells may be left, thus the disease is known as minimal residual disease (MRD), which is proven to be responsible for clinical relapse. Detection and quantification of MRD in ALL patients after chemotherapy might improve therapeutic results and prognosis.
Methods: Polymerase chain reaction (PCR) and nested-PCR were used to detect rearranged immunoglobulin heavy-chain (IgH) CDRIII gene and T lineage cell receptor (TCR) V¦Ä2D¦Ä3 gene for 71 ALL patients. Cerebral spinal fluid (CSF) samples from 67 patients at CR were also detected by the same methods. Leukemic clones were quantified by limiting dilution analysis.
Results: IgH CDRIII rearrangements were found positive in 43 (82.7%) of 52 patients with B lineage ALL and 3 (15.8%) of 19 patients with T lineage ALL. TCR V¦Ä2D¦Ä3 positive rearrangements were present in 17 (32.7%) patients with B lineage ALL and 12 (63.2%) with T lineage ALL. MRD was quantified with the nested-PCR through limiting dilution of DNA samples from these rearrangement positive patients. Patients with MRD level above 2¡Á10-3 in the first CR were found to have a high relapse rate and this situation was also found in patients with a slow decrease or a high level of MRD. Whereas patients with undetectable or detected less than 2¡Á10-5 level of MRD in the first CR had a good prognosis. Of the 67 CSF samples, 9 were positive for IgH CDRIII rearrangement and 4 positive for TCR V¦Ä2D¦Ä3 rearrangement; 5 of them eventually developed central nervous system leukemia.
Conclusions: The relationship between MRD detection and outcome is significant for ALL patients. Early quantification of leukemic cells after chemotherapy may be an effective strategy for predicting ou tcome and hence individualizing ALL treatment in childhood.
Key words: minimal residual disease; acute lymphoblastic leukemia; polymerase chain reaction; immunoglobulin heavy-chain; T lineage cell receptor; gene rearrangement
World J Pediatr 2007;3(4):290-294
Introduction
Leukemic cells can reach 1012 or higher in patients with acute lymphoblastic leukemia (ALL) at diagnosis. They will fall below 109 when the patients get complete remission (CR) after induction chemotherapy. But a few residual leukemic cells remain and are known as the minimal residual disease (MRD). With current intensive chemotherapeutic protocols, more than 95% of children with ALL can achieve CR after induction therapy, but up to 30% will ultimately relapse and most of them will die. MRD is proven to be responsible for clinical relapse. Hence it is very important to detect the MRD in ALL patients. The detection of MRD might be useful in judging curative effect, predicting early relapse and assessing prognosis.[1-3]
In this study, the relationship between MRD and the prognosis of ALL was assessed in bone marrow (BM) samples from 71 untreated children with ALL and 67 cerebral spinal fluid (CSF) samples in CR by polymerase chain reaction (PCR) or nested-PCR[4-6] using the immunoglobulin heavy-chain (IgH) CDRIII gene and T lineage cell receptor (TCR) V¦Ä2D¦Ä3 gene as target markers.[7-13] Both gene rearrangements in bone marrow samples of ALL patients after CR were quantitatively analyzed by limiting dilution assay. The results indicated that the method was highly sensitive, capable of detecting as few as 1 leukemic cell in 106 normal cells.[14]
Methods
BM samples were taken from 71 patients with ALL aged from 1 to 14 years, diagnosed at our hospital from 1999 to 2001. Among them, 45 were boys and 26 girls. According to FAB classification, 49 patients belonged to L1 type, 17 L2 and 5 L3. Immunophenotypes of the 71 patients detected by flow cytometry (FCM) were as follows: T lineage ALL (19 patients) and B lineage ALL (52). BM samples were taken for detecting the clonal rearrangements of IgH CDRIII gene and TCR V¦Ä2D¦Ä3 gene, respectively.
CSF samples were taken from 67 patients who had reached CR during 1-24 months.[15] All patients had either TCR V¦Ä2D¦Ä3 gene or IgH CDRIII gene clonal rearrangement in BM samples.
Twenty-nine patients with TCR V¦Ä2D¦Ä3 gene clonal rearrangement and 46 patients with IgH CDRIII gene clonal rearrangement were further studied for monitoring MRD at the end of induction and whenever possible, 1-35 months thereafter. After one course of standard induction therapy, all of these patients achieved CR. Similarly, DNA samples were obtained from BM samples of all patients and amplified by nested-PCR. The patients were followed up at least for 1-35 months.
BM samples from 30 children diagnosed with immune thrombocytopenic purpura (ITP) served as the control group.
DNA preparation
Mononuclear cells were isolated using Histopaque-1077 for centrifugation, and DNA was extracted using standard procedures.[16] Briefly, cells were lysed in lysis buffer with proteinase K, followed by phenol/chloroform extraction and ethanol precipitation. DNA pellets were resuspended in TE buffer and stored at -20ºC until use. For CSF samples, DNA was prepared using the simple boiling procedure.
PCR detection
DNA samples were screened by PCR for clonal IgH gene rearrangement using consensus primers VH and JH, and for TCR V¦Ä2D¦Ä3 gene rearrangement using consensus pairs of outer and inner primers of V¦Ä2 and D¦Ä3. Sequences of oligonucleotide primers are listed in the Table.
Amplification for IgH CDRIII gene rearrangement was performed by an initial denaturation at 94ºC, followed by 35 cycles at 94ºC for 30 seconds, 55ºC for 30 seconds, and 72ºC for 60 seconds, and a final extension at 72ºC for 7 minutes. Amplification for TCR V¦Ä2D¦Ä3 gene rearrangement was performed using nested-PCR. The first step was conducted using outer primers at 94ºC for 2 minutes, 56ºC for 2 minutes, and 72ºC for 3 minutes in 35 cycles and final extension at 72ºC for 7 minutes. This step was followed by the second amplification using PCR products as the templates and the inner primers in 35 cycles at 94ºC for 45 seconds, 56ºC for 1 minute, 72ºC for 2 minutes and final extension at 72ºC for 7 minutes in a volume of 25 ¦Ìl of the reaction mixture containing 1 ¦Ìg DNA, 25 pmol of each primer, 200 ¦Ìmol/L dNTP, 1 U TaqDNA polymerase and 50 mmol/L MgCL2. PCR products were analyzed through electrophoresis with 2.5% agarose gel.
A series of 10-fold dilutions were made from each DNA sample, and added to PCR reaction[17] to give templates consisting of 0.0001 to 500 000 diploid genomes (1 cell contains 6 pg DNA). The sensitivity of the limiting dilution assay was assessed for each patient-specific TCR V¦Ä2D¦Ä3 gene rearrangement.
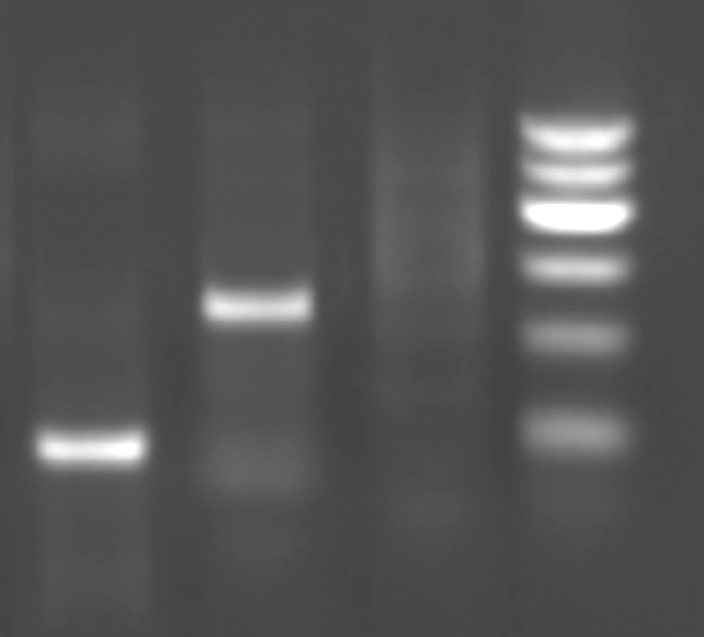
Fig. A: marker; B: control (ITP patients); C: IgH CDRIII gene rearrangement; D: TCR V¦Ä2D¦Ä3 gene rearrangement.
Calculation of MRD
Each patient-specific sensitivity = DNA level (pg) in PCR reactions when amplification was negative ¡Á the percentage of primitive lymphoblastic cell in BM/6. The level of MRD = limiting dilution fold ¡Á the patient's sensitivity / the amount of nucleated cells.[18]
Statistical analysis
The relationship between MRD and the outcome was analyzed in accordance with P by obtaining geometric mean using Wilcoxon rank-sum test.
Results
The characteristic patterns of IgH CDRIII and TCRV ¦Ä2D¦Ä3 gene clonal rearrangements from 71 patients with ALL on initial therapy are shown in the Fig. One band was presented at 110-130 bp in positive clonal IgH CDRIII gene rearrangement and at 80-110 bp in TCR V¦Ä2D¦Ä3 gene clonal rearrangement. No band could be observed in DNA samples from 30 ITP patients. Positive IgH CDRIII rearrangement was found in 43 (82.7%) of 52 patients with B lineage ALL and 3 (15.8%) of 19 patients with T lineage ALL. TCR V¦Ä2D¦Ä3 positive rearrangements were present in 17 (32.7%) B lineage ALL and 12 (63.2%) T lineage ALL. Sixty-three patients (88.7%) were found to be positive for both gene rearrangements.
MRD levels were quantified in 29 patients with TCR V¦Ä2D¦Ä3 gene clonal rearrangement and 46 patients with IgH CDRIII gene clonal rearrangement. MRD levels were monitored in ALL patients with clonal TCR V¦Ä2D¦Ä3 and IgH CDRIII gene rearrangements at the time of first CR after induction therapy[19,20] and during the period of 1-35 months thereafter. Multiple MRD was detected in these patients during the treatment and follow-up.
MRD was not found in 14 (48.3%) of 29 TCR V¦Ä2D¦Ä3 detected patients after reaching CR at the end of induction therapy. In the following chemotherapy, 12 patients were MRD-negative and showed clinical CR, but the other 2 patients reversed from MRD negative to positive, and presented central nervous system leukemia (CNSL). However, with intensive chemotherapy, the MRD level fell down to negative until reaching clinical CR. At the time when this paper was published, the 2 patients still had no signs of relapse. The remaining 15 (51.7%) TCR V¦Ä2D¦Ä3 detected patients were MRD positive at the end of induction therapy. These patients were divided into three groups according to their MRD levels. The first group included 4 patients with high MRD levels (¡Ý10-3) when reaching the first CR. After chemotherapy, 1 patient kept a high level of MRD and the other 3 patients showed slowly decreased MRD level. In this group, 2 (50%) patients relapsed after 5-7 months of follow-up, and the other 2 are still in relief. The second group comprised 7 patients with intermediate MRD levels (2¡Á10-5-10-3). Five of them had a rapid decrease of MRD levels and achieved a steady CR, but the other 2 (28.6%) patients with a slow decrease of MRD levels relapsed in 6 months after the first CR. The third group consisted of 4 patients with low MRD levels (<2¡Á10-5) at the end of induction therapy, and their MRD levels quickly became negative. These patients achieved steady clinical CR. Among the 29 patients, 4 (13.8%) relapsed, including 2 patients of high-risk subtype, and their median MRD level was 1.875¡Á10-3. By contrast, 25 (86.2%) patients with a steady CR had a median MRD level of 3¡Á10-6 (P<0.01). In the patients with high-risk ALL, MRD levels were significantly higher in the relapse group than in the non-relapse group (P<0.05).
Similar results were found in the 46 patients with IgH CDRIII gene clonal rearrangement. Twenty-two (47.8%) of the 46 patients showed no MRD after reaching CR by the end of induction therapy. During the continued chemotherapy, 18 patients were persistently negative for MRD and had clinical CR, but the other 4 patients had MRD levels reversed from negative to positive. Then the MRD levels fell down to negative again after intensive chemotherapy. In the remaining 24 (52.3%) patients, MRD was still detectable by the end of induction therapy. These patients could be divided into three groups according to the TCR V¦Ä2D¦Ä3 gene clonal rearrangement. The first group included 7 patients with high MRD levels (¡Ý10-3) after the first CR. MRD levels remained high in 3 patients, but decreased slowly in the other 4 patients. In this group, 4 (57.1%) patients relapsed during a follow-up of 9-13 months. The second group consisted of 11 patients with intermediate MRD levels (2¡Á10-5-10-3). Eight patients experienced rapid decrease of their MRD levels, and the other three patients had a slow decrease in their MRD levels. Two (18.2%) patients relapsed at a period of 15-18 months after the first CR. The third group had 6 patients with a low MRD level (<2¡Á10-5). The MRD levels in all the 6 patients were quickly reversed to negative and a steady CR was achieved. Among the 46 patients, 6 (13.1%) including 5 patients with high-risk ALL relapsed, and their MRD median level was 2.1¡Á10-3. By contrast, 40 (86.9%) patients with a steady CR had a median MRD level of 5.2¡Á10-6, which was statistically significant as compared to that of the relapse group (P<0.01).
TCR V¦Ä2D¦Ä3 gene clonal rearrangement and IgH CDRIII gene clonal rearrangement were also detected in the CSF samples from the 67 patients with either TCR V¦Ä2D¦Ä3-positive or IgH CDRIII-positive diagnosed from BM specimens. The results showed that 9 patients were positive in IgH CDRIII rearrangement and 4 positive in TCR V¦Ä2D¦Ä3 rearrangement. All the rearrangemental types detected were the same as in BM samples while routine examinations of CSF were normal with no leukemic cells found. Among them, 10 patients received more intensive intrathecal treatment (IT) by triple chemotherapy including methotrexate, cytarabine and dexamethasone in order to control CNSL. However, 2 of the 10 patients who received intensive IT therapy and another 3 patients who refused to do it developed CNSL. The results indicated that dynamic detection of IgH and TCR gene rearrangements in CSF of ALL patients was an effective approach for early clinical diagnosis of CNSL.
Discussion
Adapted protocols for risk-group and more intensive chemotherapy have achieved a high cure rate in pediatric ALL patients, but a significant proportion will relapse afterwards. Because MRD is the source of relapse, clinicians have to monitor MRD levels effectively and to incorporate MRD data into clinical practice. Currently, the most useful MRD assays available are PCR amplification of rearranged IgH and TCR gene, and FCM detection of aberrant immunophenotypes.[21,22] PCR-based techniques are more sensitive and cost-effective than FCM.[23] And rearrangements are considered to be excellent patient-specific PCR targets for MRD detection except that these rearrangements may be unstable because of clonal evolution during the course of the disease.[24,25] Among the 71 ALL patients studied in our study, positive IgH CDRIII rearrangements were found in 43 (82.7%) of 52 patients with B lineage ALL and 3 (15.8%) of 19 with T lineage ALL. TCR V¦Ä2D¦Ä3 positive rearrangements were present in 17 (32.7%) patients with B lineage ALL and 12 (63.2%) patients with T lineage ALL. Both gene rearrangements were found in 63 (88.7%) patients. No rearrangement evolution was found in this cohort of patients perhaps because gene rearrangements of most of these patients kept stable during the disease.[26] Our results further indicate that IgH CDRIII and TCR V¦Ä2D¦Ä3 gene clonal rearrangements are useful molecular markers for detecting MRD of ALL clones.
We found that detection and quantification of MRD are closely related to prognosis. There is a striking relationship between the extent of MRD and the outcome, which could predict relapse of the disease in patients with a high level of MRD of more than 10-3 at the time of the first CR.[27] This finding is consistent with that of other groups, obviously the MRD value below 10-4 at the end of induction therapy predicts good prognosis.[28] Similarly, the level of MRD may decrease slowly or remain high in the subsequent therapy. On the other hand, the level of MRD reversing to positive or increasing gradually is also a high-risk factor for relapse. In our study, the simple detection of MRD was limited in predicting relapse. Quantitation of MRD is strongly recommended while keeping monitoring of the dynamic levels of MRD during the subsequent therapy.[29,30]
It is also a measurement for drug sensitivity and the number of leukemic cells being eradicated. This ensures early modification of therapeutic strategies. Possibly patients with a high MRD level at the first remission should receive an early and strong intensive therapy. We suggest that patients with MRD above 10-3, perhaps even above 2¡Á10-5, should receive intensive therapy. During the first 6 months of chemotherapy, patients with a high level of MRD or a slowly decreasing level should also receive intensive treatment. But those patients with MRD level below 2¡Á10-5 or undetectable at the start of consolidation treatment might be treated with less intensive therapy than that used currently in order to reduce the long-term side effect of drugs and the cost of treatment. Also PCR in monitoring the MRD of CSF is effective to detect CNSL early before the appearance of clinical symptoms.
Acknowledgements
The authors appreciate the support from all the pediatricians of the Department of Pediatrics, Union Hospital, Huazhong University of Science and Technology.
Funding: None.
Ethical approval: Not needed.
Competing interest: None declared.
Contributors: BY wrote the main body of the article under the supervision of JRM and FHB. All authors contributed to the design and interpretation of the study. JRM is the guarantor.
References
1 Bastida Vil¨¢ P, Palacio Garc¨ªa C, Solsona Riera M, Ortega Aramburu JJ, S¨¢nchez de Toledo Codina J. Minimal residual disease in acute lymphoblastic leukemia: a new concept of complete remission. An Pediatr (Barc) 2005;63:390-395.
2 Szczepanski T. Why and how to quantify minimal residual disease in acute lymphoblastic leukemia. Leukemia 2007;21:622-626.
3 Moppett J, Burke GA, Steward CG, Oakhill A, Goulden NJ. The clinical relevance of detection of minimal residual disease in childhood acute lymphoblastic leukaemia. J Clin Pathol 2003;56:249-535.
4 Campana D. Minimal residual disease studies in acute leukemia. Am J Clin Pathol 2004;122 Suppl:S47-57.
5 He YY, Ye TZ. Detection of minimal residual disease in children leukemia patients by using PCR. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2007;15:652-656.
6 Kwok CS, Kham SK, Dolendo MC, Ariffin H, Lin HP, Quah TC, et al. Molecular monitoring of minimal residual disease in childhood acute lymphoblastic leukaemia using antigen receptor gene rearrangements is highly feasible for disease stratification and prognostication. Ann Acad Med Singapore 2003;32(5 Suppl):S31-33.
7 van der Velden VH, de Bie M, van Wering ER, van Dongen JJ. Immunoglobulin light chain gene rearrangements in precursor-B-acute lymphoblastic leukemia: characteristics and applicability for the detection of minimal residual disease. Haematologica 2006;91:679-682.
8 Sudhakar N, Nancy NK, Rajalekshmy KR, Ramanan G, Rajkumar T. T-cell receptor gamma and delta gene rearrangements and junctional region characteristics in south Indian patients with T-cell acute lymphoblastic leukemia. Am J Hematol 2007;82:215-221.
9 Li A, Zhou J, Zuckerman D, Rue M, Dalton V, Lyons C, et al. Sequence analysis of clonal immunoglobulin and T-cell receptor gene rearrangements in children with acute lymphoblastic leukemia at diagnosis and at relapse: implications for pathogenesis and for the clinical utility of PCR-based methods of minimal residual disease detection. Blood 2003;102:4520-4526.
10 Donovan JW, Ladetto M, Zou G, Neuberg D, Poor C, Bowers D, et al. Immunoglobulin heavy-chain consensus probes for real-time PCR quantification of residual disease in acute lymphoblastic leukemia. Blood 2000;95:2651-2658.
11 Hoshino A, Funato T, Munakata Y, Ishii T, Abe S, Ishizawa K, et al. Detection of clone-specific immunoglobulin heavy chain genes in the bone marrow of B-cell-lineage lymphoma after treatment. Tohoku J Exp Med 2004;203:155-164.
12 Bonjean B, Grollet L, Visentin E, Sigaux F, Cayuela JM. Development of a new strategy for minimal residual disease monitoring in children with B-precursor acute lymphoblastic leukemia. Ann Biol Clin (Paris) 2004;62:465-470.
13 Br¨¹ggemann M, van der Velden VH, Raff T, Droese J, Ritgen M, Pott C, et al. Rearranged T-cell receptor beta genes represent powerful targets for quantification of minimal residual disease in childhood and adult T-cell acute lymphoblastic leukemia. Leukemia 2004;18:709-719.
14 Dawidowska M, Wachowiak J, Witt M. Molecular methods for diagnostics and assessment of treatment effectiveness in modern pediatric hematooncology. Postepy Biochem 2006;52:408-416.
15 Nuñez CA, Zipf TF, Roberts WM, Medeiros LJ, Hayes K, Bueso-Ramos CE. Molecular monitoring of cerebrospinal fluid can predict clinical relapse in acute lymphoblastic leukemia with eosinophilia. Arch Pathol Lab Med 2003;127:601-605.
16 Sambbrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold spring habour laboratory. NY: Cold Spring Habour Laboratory Presss, 1989: 464.
17 Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992;13:444-449.
18 Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantification of targets for PCR by use of limiting dilution. Biotechniques 1992;13:444-449.
19 Okamoto T, Yokota S, Katano N, Seriu T, Nakao M, Taniwaki M, et al. Minimal residual disease in early phase of chemotherapy reflects poor outcome in children with acute lymphoblastic leukemia¡ªa retrospective study by the Children's Cancer and Leukemia Study Group in Japan. Leuk Lymphoma 2002;43:1001-1006.
20 Marshall GM, Haber M, Kwan E, Zhu L, Ferrara D, Xue C, et al. Importance of minimal residual disease testing during the second year of therapy for children with acute lymphoblastic leukemia. J Clin Oncol 2003;21:704-709.
21 Settin A, Al Haggar M, Al Dosoky T, Al Baz R, Abdelrazik N, Fouda M, et al. Prognostic cytogenetic markers in childhood acute lymphoblastic leukemia. Indian J Pediatr 2007;74:255-263.
22 Zhou J, Goldwasser MA, Li A, Dahlberg SE, Neuberg D, Wang H, et al. Quantitative analysis of minimal residual disease predicts relapse in children with B-lineage acute lymphoblastic leukemia in DFCI ALL Consortium Protocol 95-01. Blood 2007;110:1607-1611.
23 Gameiro P, Moreira I, Yetgin S, Papaioannou M, Potter MN, Prentice HG, et al. Polymerase chain reaction (PCR)- and reverse transcription PCR-based minimal residual disease detection in long-term follow-up of childhood acute lymphoblastic leukaemia. Br J Haematol 2002;119:685-696.
24 Germano G, del Giudice L, Palatron S, Giarin E, Cazzaniga G, Biondi A, et al. Clonality profile in relapsed precursor-B-ALL children by GeneScan and sequencing analyses. Consequences on minimal residual disease monitoring. Leukemia 2003;17:1573-1582.
25 Guggemos A, Eckert C, Szczepanski T, Hanel C, Taube T, van der Velden VH, et al. Assessment of clonal stability of minimal residual disease targets between 1st and 2nd relapse of childhood precursor B-cell acute lymphoblastic leukemia. Haematologica 2003;88:736-746.
26 Li AH, Forestier E, Rosenquist R, Roos G. Minimal residual disease quantification in childhood acute lymphoblastic leukemia by real-time polymerase chain reaction using the SYBR green dye. Exp Hematol 2002;30:1170-1177.
27 Pui CH, Campana D. New definition of remission in childhood acute lymphoblastic leukemia. Leukemia 2000;14:783-785.
28 Settin A, Al Haggar M, Al Dosoky T, Al Baz R, Abdelrazik N, Fouda M, et al. Prognostic cytogenetic markers in childhood acute lymphoblastic leukemia: cases from Mansoura, Egypt. Hematology 2006;11:341-349.
29 Mortuza FY, Papaioannou M, Moreira IM, Coyle LA, Gameiro P, Gandini D, et al. Minimal residual disease tests provide an independent predictor of clinical outcome in adult acute lymphoblastic leukemia. J Clin Oncol 2002;20:1094-1104.
30 Lo-Coco F, Ammatuna E. Front line clinical trials and minimal residual disease monitoring in acute promyelocytic leukemia. Curr Top Microbiol Immunol 2007;313:145-156.
Received July 7, 2006 Accepted after revision September 20, 2007
|

